Current Software
GitHub
We have moved our software development to GitHub. Find our latest software there
Medulloblastoma Data Explorer
The Medulloblastoma Data Explorer is an interactive data explorer of medulloblastoma proteomic and phosphoproteomic (plus other ‘omics) data sets. It is the companion app of our medulloblastoma proteomics paper (Archer, Ehrenberger, Mundt, et al. Cancer Cell (2018)).
Past Software
Omics Integrator
Omics Integrator is package comprised of command-line tools designed to integrate high-throughput datasets such as gene expression, phospho-proteomic data and the results from genetic screens. As shown below, Garnet is used to identify transcription factors that give rise to gene expression changes using epigenetic data while Forest integrates these data or other data by finding connections in a protein interaction network.
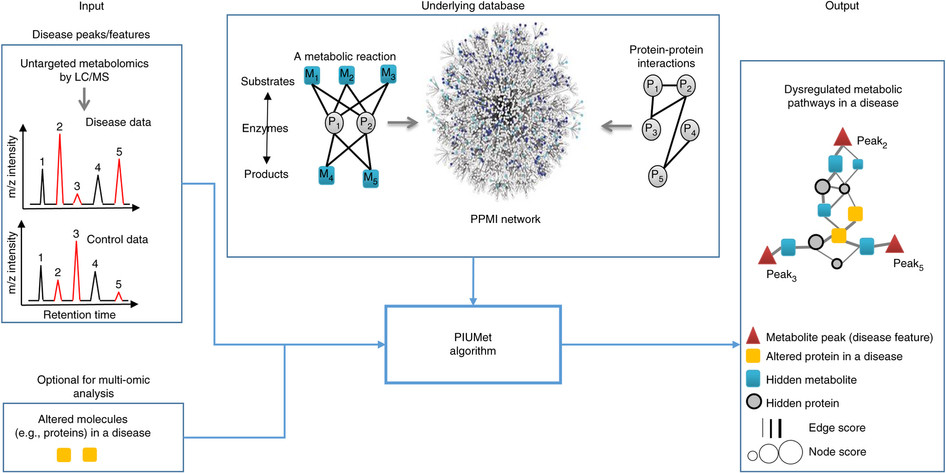
PIUMet
PIUMet is a network-based algorithm for integrative analysis of untargeted metabolomic data. It leverages known metabolic reactions and protein-protein interactions to analyze the ambiguous assignment of metabolomics features and identify disease-associated pathways and hidden components.
SAMNet
SAMNet uses a constrained optimization approach to identify relationships between high-throughput mRNA expression data with other measured changes (i.e. genetic hits, phosphoproteomics). The result is a compact network with edges representing protein interactions that best explain the changes in mRNA expression dowstream of the upstream changes for each condition.
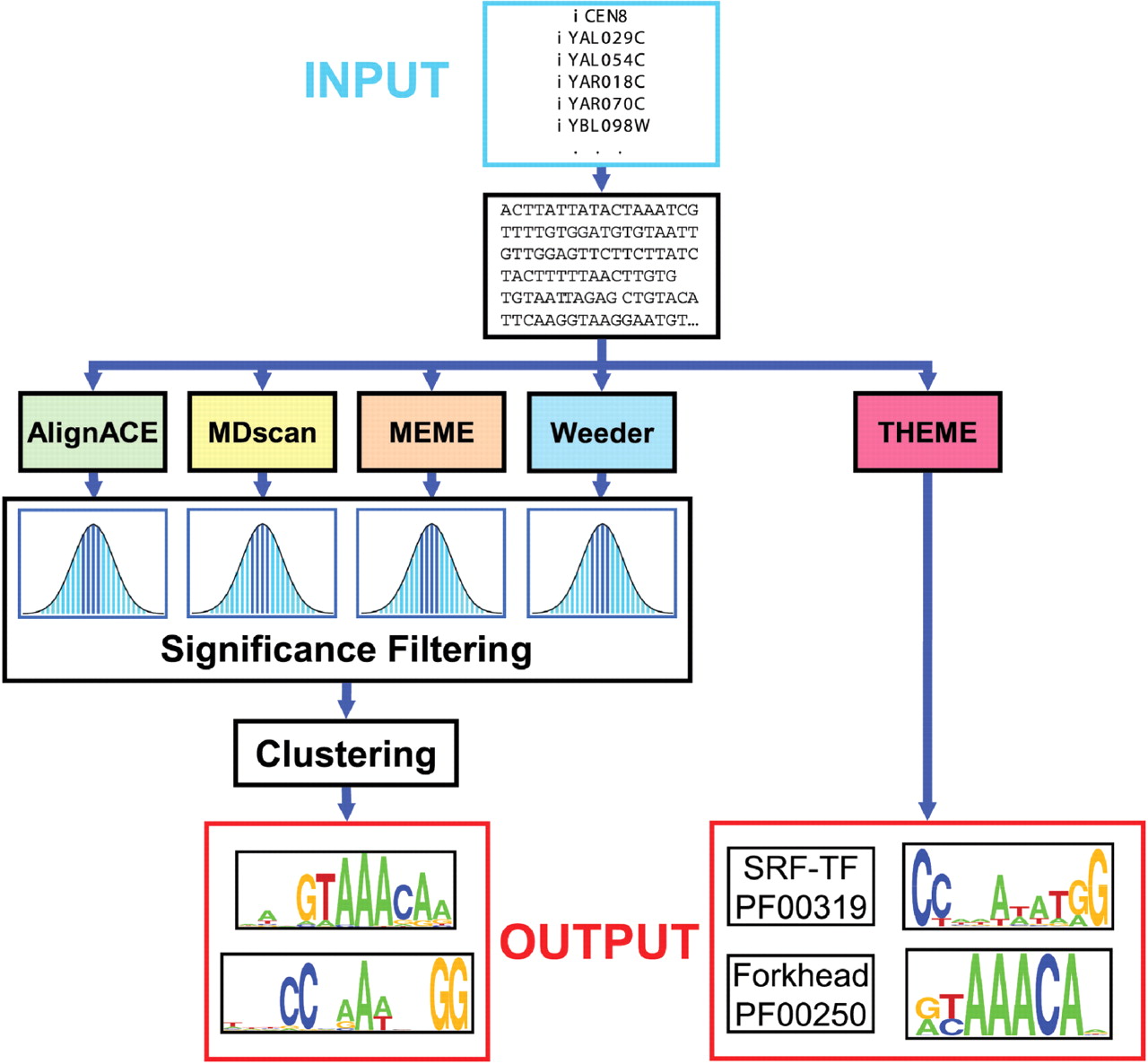
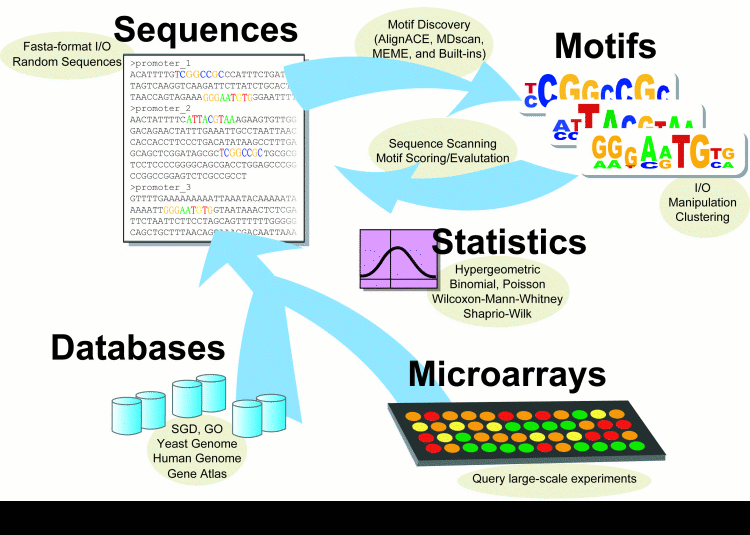
Webmotifs
WebMOTIFS is an online tool for motif discovery, scoring, analysis, and visualization. It allows you to use different programs to search for DNA-sequence motifs, and to easily combine and evaluate the results.
TAMO
Motif analysis application for 3T3-L1 DNase hypersensitivity data.
AdipoSight
Motif analysis application for 3T3-L1 DNase hypersensitivity data.
ResponseNet
Revealing signaling and regulatory networks linking genetic and transcriptomic screening data
An Improved Map of Conserved Regulatory Sites for Saccharomyces cerevisiae
The regulatory map of a genome consists of the binding sites for proteins that determine the transcription of nearby genes. An initial regulatory map for S. cerevisiae was recently published using six motif discovery programs to analyze genome-wide chromatin immunoprecipitation data for 203 transcription factors. The programs were used to identify sequence motifs that were likely to correspond to the DNA-binding specificity of the immunoprecipitated proteins. We report improved versions of two conservation-based motif discovery algorithms, PhyloCon and Converge. Using these programs, we create a refined regulatory map for S. cerevisiae by reanalyzing the same chromatin immunoprecipitation data.